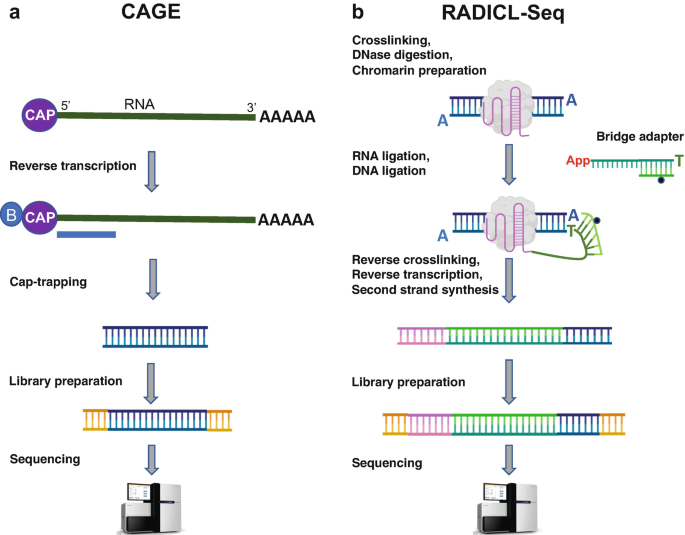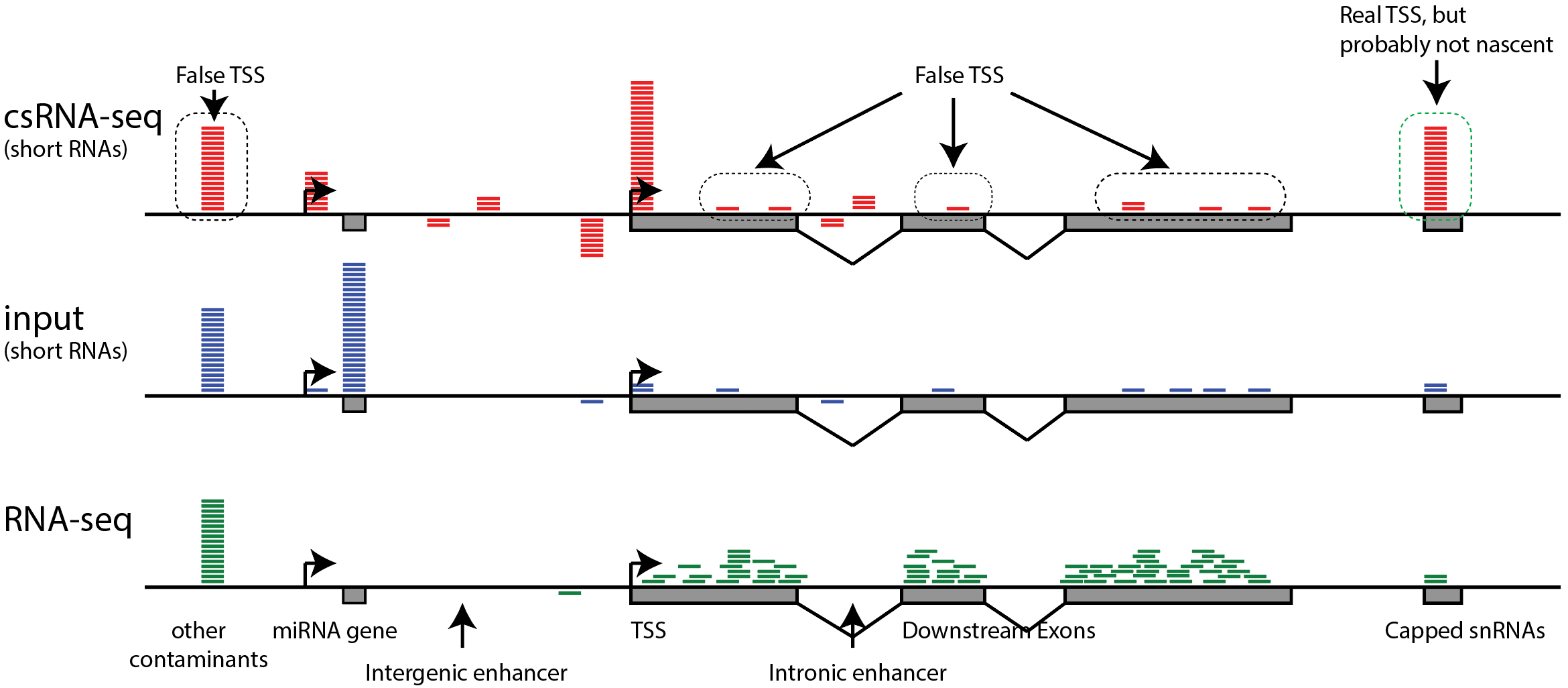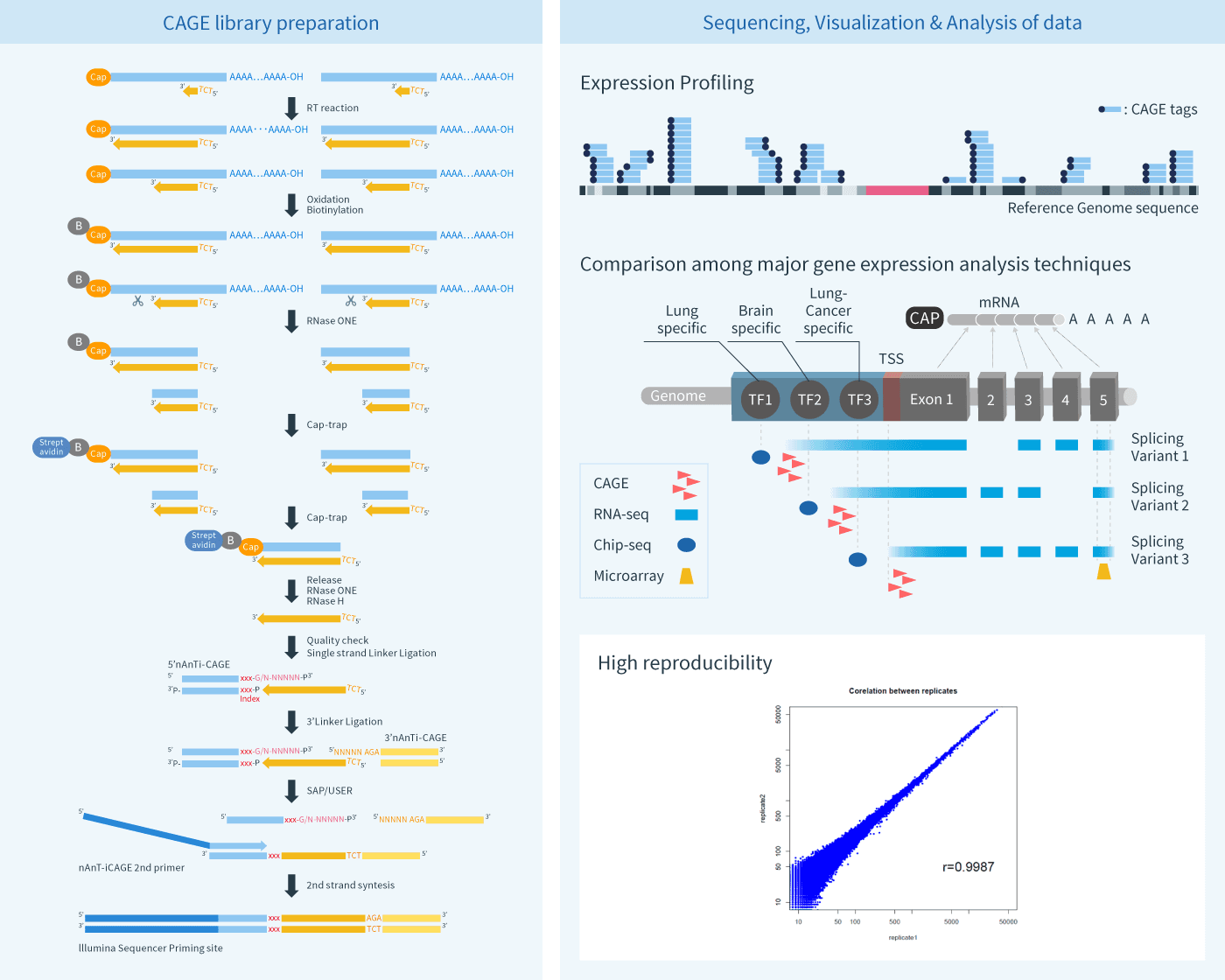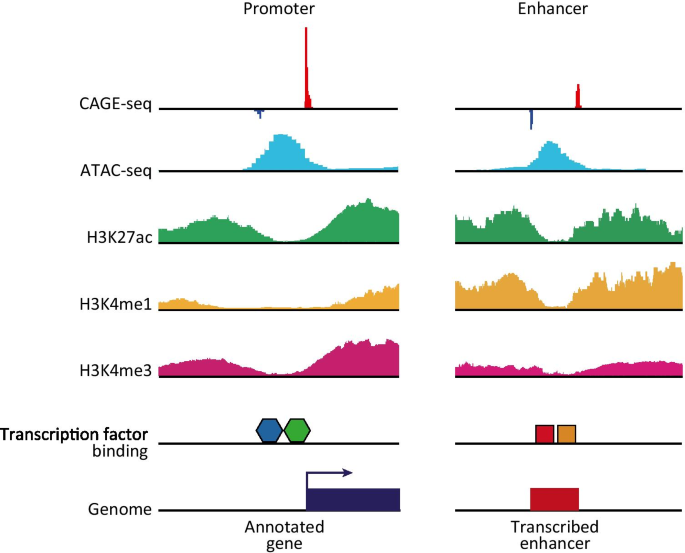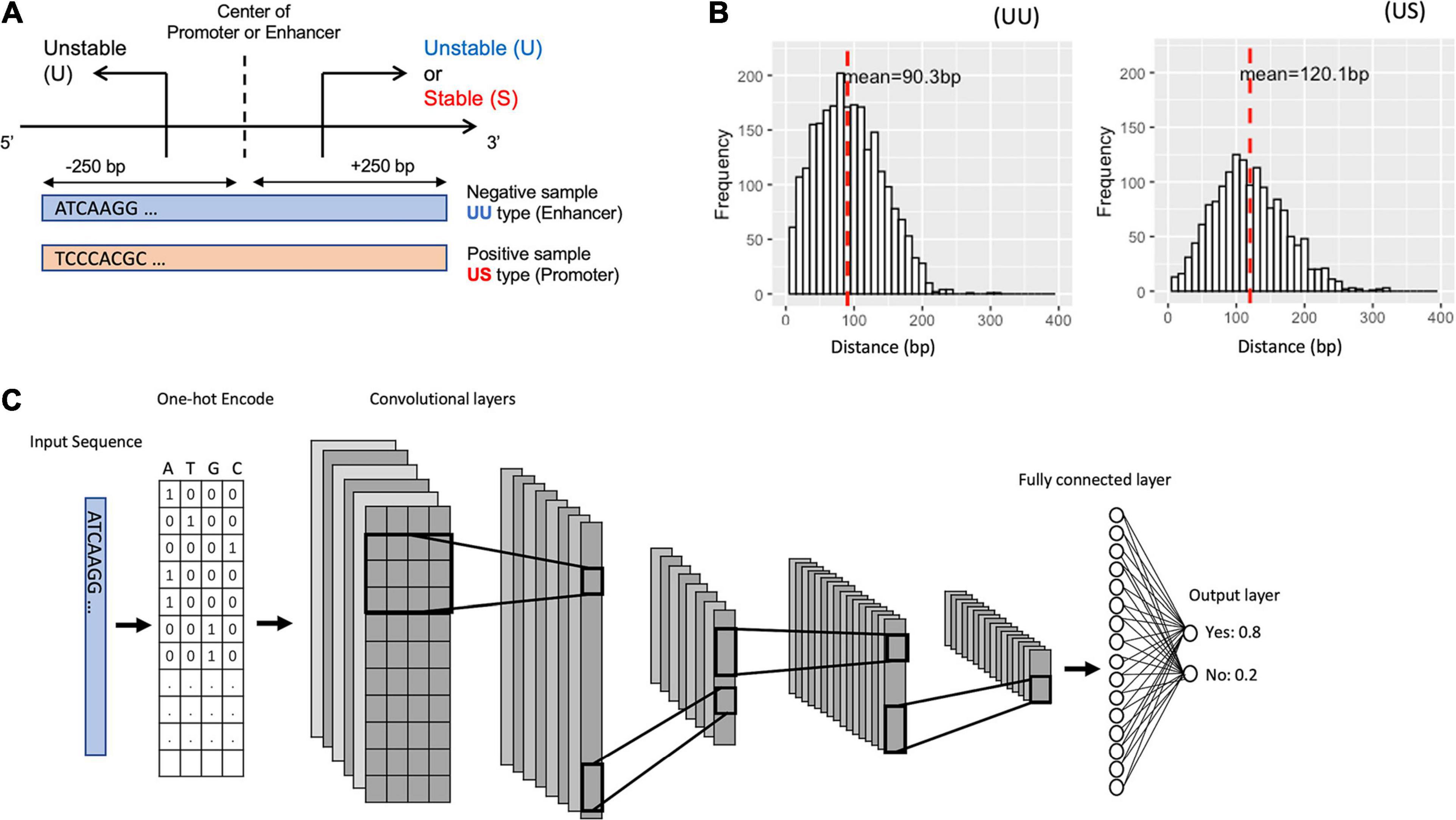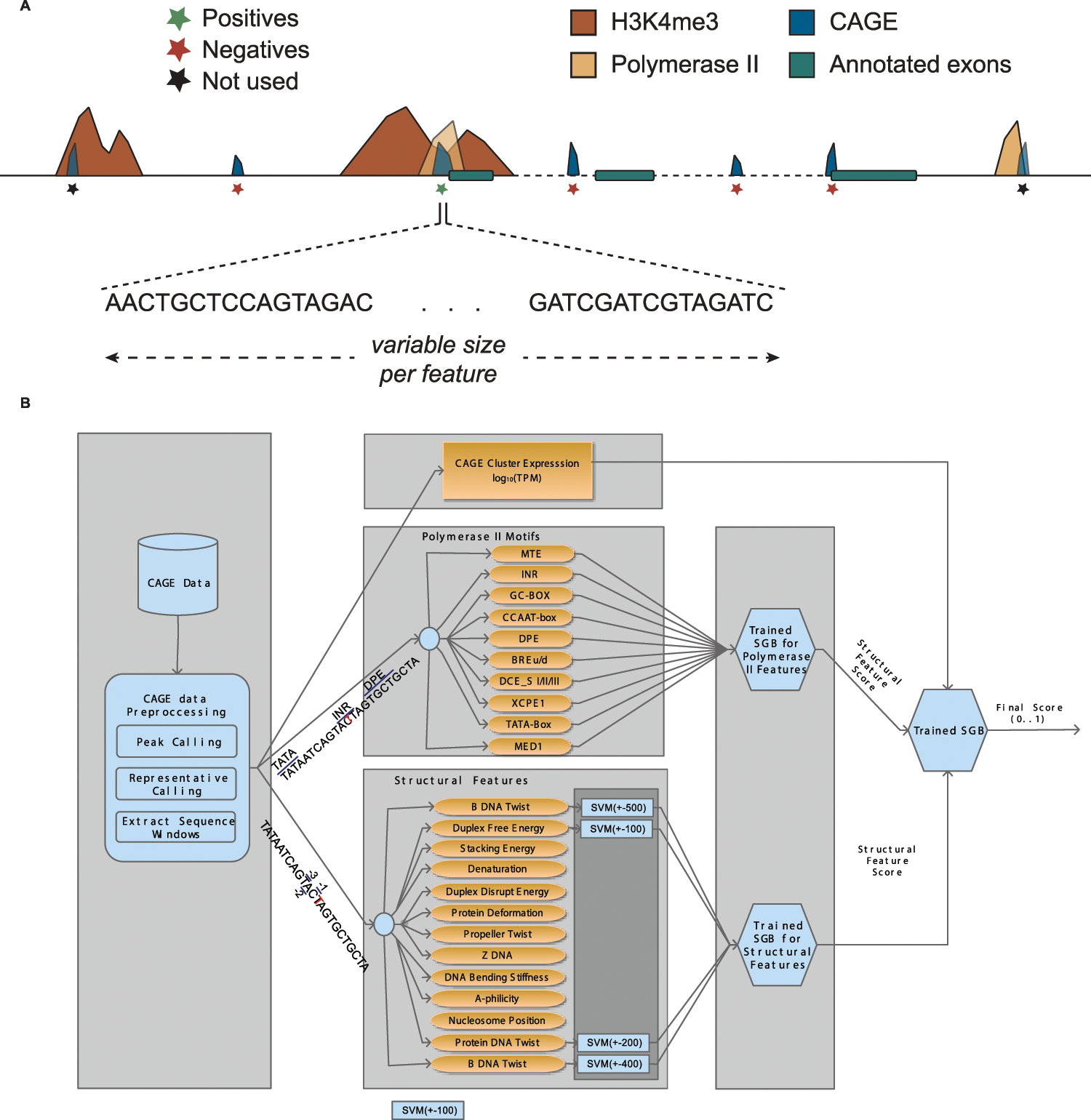
Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data | Scientific Reports
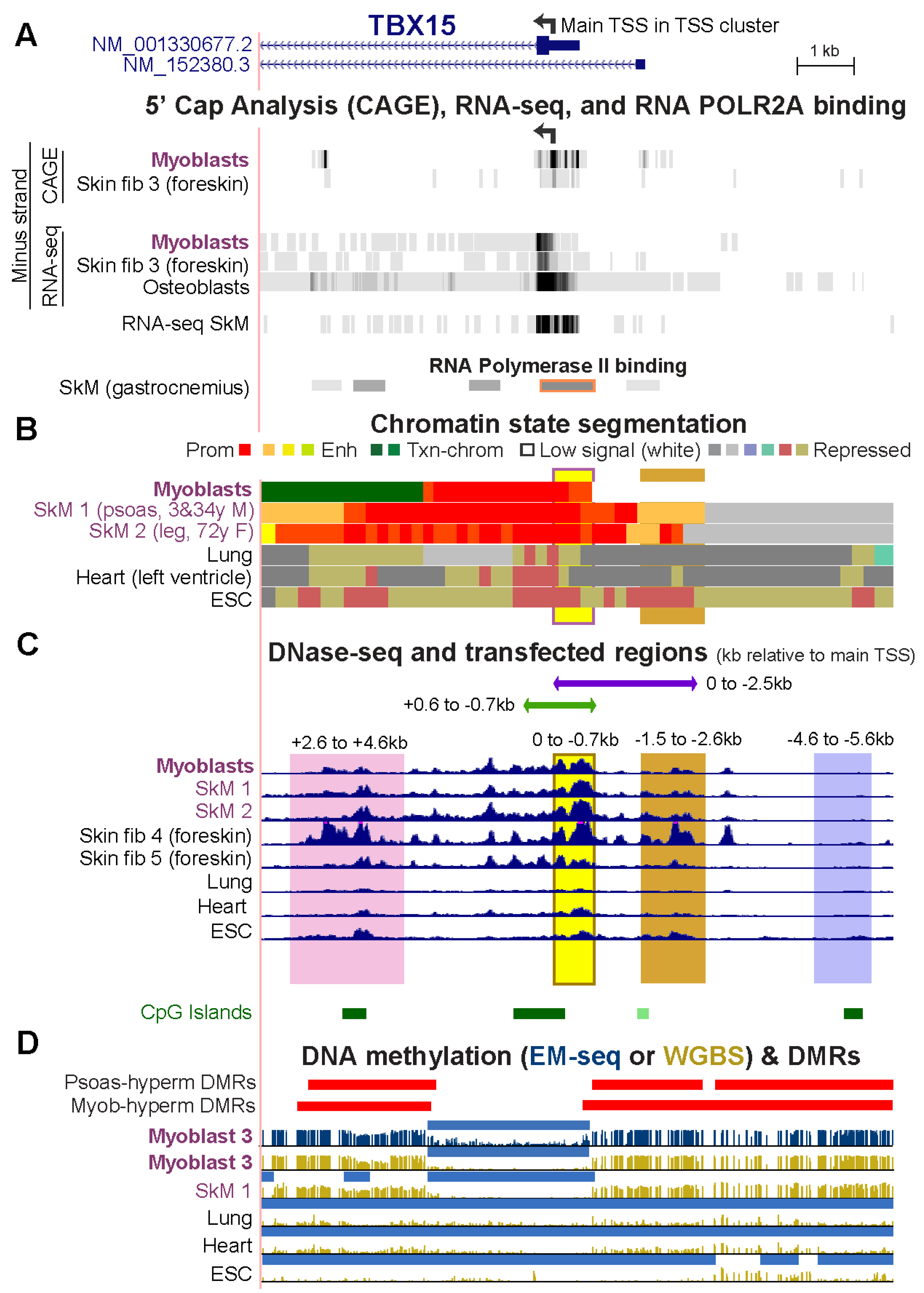
Epigenomes | Free Full-Text | Promoter-Adjacent DNA Hypermethylation Can Downmodulate Gene Expression: TBX15 in the Muscle Lineage

NET-CAGE characterizes the dynamics and topology of human transcribed cis-regulatory elements | Nature Genetics

Cap analysis of gene expression (CAGE) sequencing reveals alternative promoter usage in complex disease | bioRxiv
Influence of Rotational Nucleosome Positioning on Transcription Start Site Selection in Animal Promoters | PLOS Computational Biology
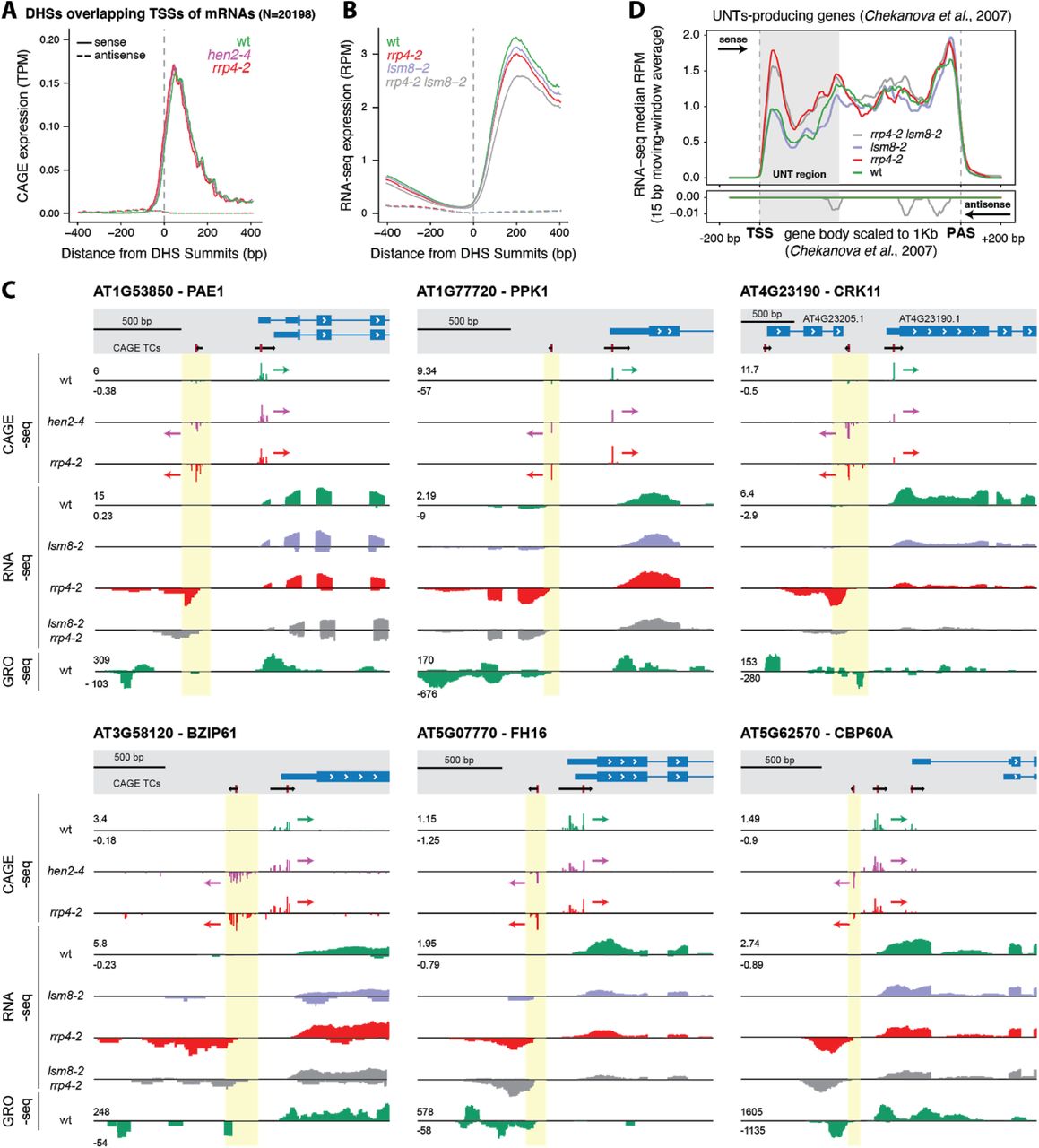
Characterization of Arabidopsis thaliana promoter bidirectionality and antisense RNAs by depletion of nuclear RNA decay enzymes | bioRxiv

Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for analysis of CAGE data | bioRxiv
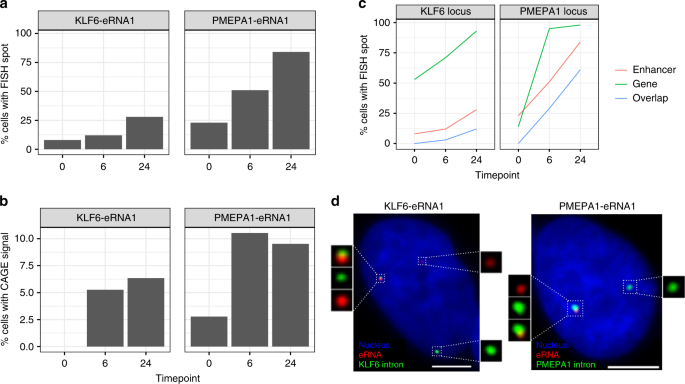
C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution | Nature Communications
Distal CpG islands can serve as alternative promoters to transcribe genes with silenced proximal promoters

Promoter sequence and architecture determine expression variability and confer robustness to genetic variants | bioRxiv